Interactive tools for visualisation of DNA, RNA, and protein structures
InfraVis Collaborators
MAX IV
InfraVis Application Expert
Joakim Bohlin (Chalmers)
InfraVis Node Coordinator
Emanuel Larsson (LU), Fabio Latino (Chalmers)
Tools & Skills
VR, AR, PDB
Keywords
Nanotechnology, Web, Augmented Reality, Protein Data Base
Background
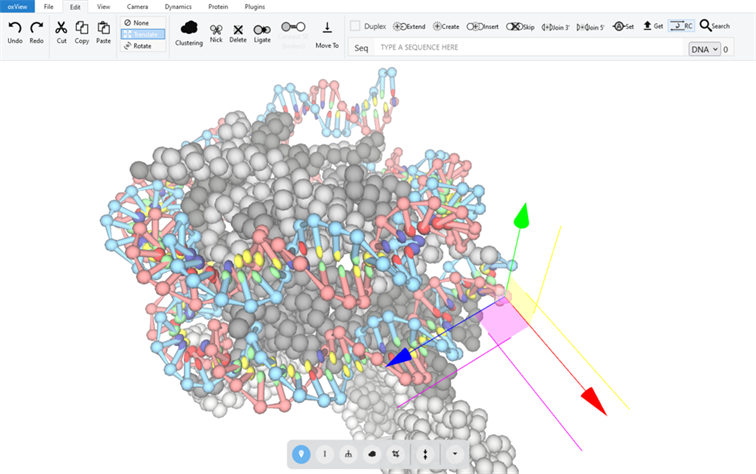
As part of his DPhil (PhD) at Oxford University, in collaboration with colleagues at Arizona State University, Joakim Bohlin developed oxView, a web-based application to design, edit, simulate, and analyse DNA, RNA, and protein structures.
Now, as an application expert at InfraVis, Joakim has used his knowledge to develop immersive web applications for visualizing protein structures (Protein Data Based – PDB – files) characterized through MAX IV beamlines in augmented reality (AR).
The results were presented at the MAX IV Users’ Meeting 2025 in Lund, Sweden. All three web applications are webXR enabled, meaning that any user with a VR headset can navigate to a web site in the device browser, inside the VR headset, and launch an immersive experience.
OxView
The main application was developed outside of InfraVis. It has seen widespread adoption within the field of DNA nanotechnology, specifically for DNA origami designs.

For more information, see:
- Bohlin, J., Matthies, M., Poppleton, E., Procyk, J., Mallya, A., Yan, H., & Šulc, P. (2022). Design and simulation of DNA, RNA and hybrid protein–nucleic acid nanostructures with oxView. Nature protocols, 17(8), 1762-1788.
- Poppleton, E., Bohlin, J., Matthies, M., Sharma, S., Zhang, F., & Šulc, P. (2020). Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation.Nucleic acids research, 48(12), e72-e72.
If any researcher is interested in InfraVis user support related to oxView, feel free to contact InfraVis (or Joakim directly).
Application: https://oxview.org
Source code: https://github.com/sulcgroup/oxdna-viewer/

OxJenga
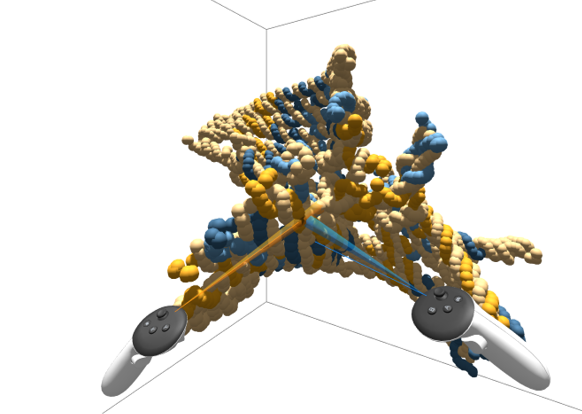
While oxView has basic VR capabilities (there is a webVR button in the View menu), an AR (Augumented Reality) visualisation was initially developed by Michael Matthies (now at Technical University of Munich, Germany).
As an initial collaboration between the Swedish research infrastructures InfraVis and MAX IV, Joakim Bohlin extended the oxJenga application to include live simulation data (from an oxDNA server) and using VR controllers.
Using a standalone headset such as the Meta Quest, a researcher can manipulate a DNA origami structure by placing external forces, seeing the Molecular Dynamics simulation update in real time. This is useful for moving the structure between different energy minimums, thereby exploring different states that may appear during lab experiments.
Application: https://akodiat.github.io/vrMaxIV/
Source code: https://github.com/Akodiat/vrMaxIV

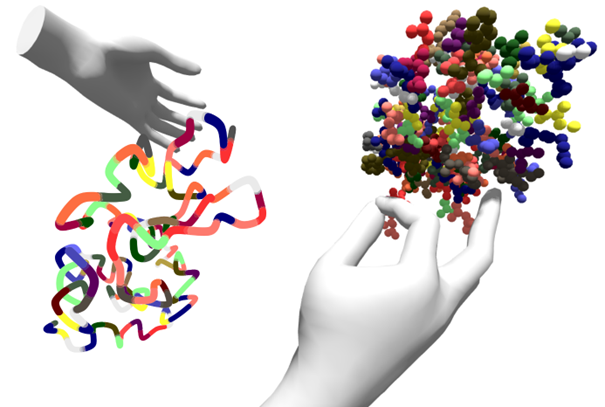
Vrotein
The third web application, Vrotein (a word game on VR and proteins), was developed entirely within InfraVis, with the purpose of being showcased at the MAX IV Users’ Meeting in January 2025.
Any protein can be loaded directly into the web app; the user only needs to input the PDB ID of the protein. The web app runs a simplified spring network model to simulate molecular dynamics, allowing the user to push and pull the amino acids with their hands (no need for separate controllers). The protein size is limited by the processing power of the headset, but future versions could include a separate simulation backend (as was done for oxJenga).
Furthermore, the structures can be visualized using different representations, such as atomic spheres, amino acid spheres, lines and splines.

Application: https://akodiat.github.io/vrotein/
Source code: https://github.com/Akodiat/vrotein
